February 3, 2021 (Huntsville, Ala.) – Researchers at the HudsonAlpha Institute for Biotechnology used a new, cutting-edge genomic sequencing technology to help physicians make diagnoses for two pediatric patients who had been on long diagnostic journeys.
Limitations of traditional sequencing in neurodevelopmental disease diagnosis
Neurodevelopmental diseases, many of which are genetic in nature, affect one to three percent of children and cause a range of physical and intellectual disabilities. Identifying the genetic variants, or changes in DNA, that lead to these diseases can provide a precise diagnosis, guide treatment approaches, and give families the answer to their years-long medical mystery.
Despite advances in genome sequencing technology used in the diagnosis of many genetic disorders, specific diagnoses for children with neurodevelopmental diseases remain elusive in many cases . This is likely because many disease-causing genetic variants are difficult or impossible to detect through typical genomic sequencing approaches.
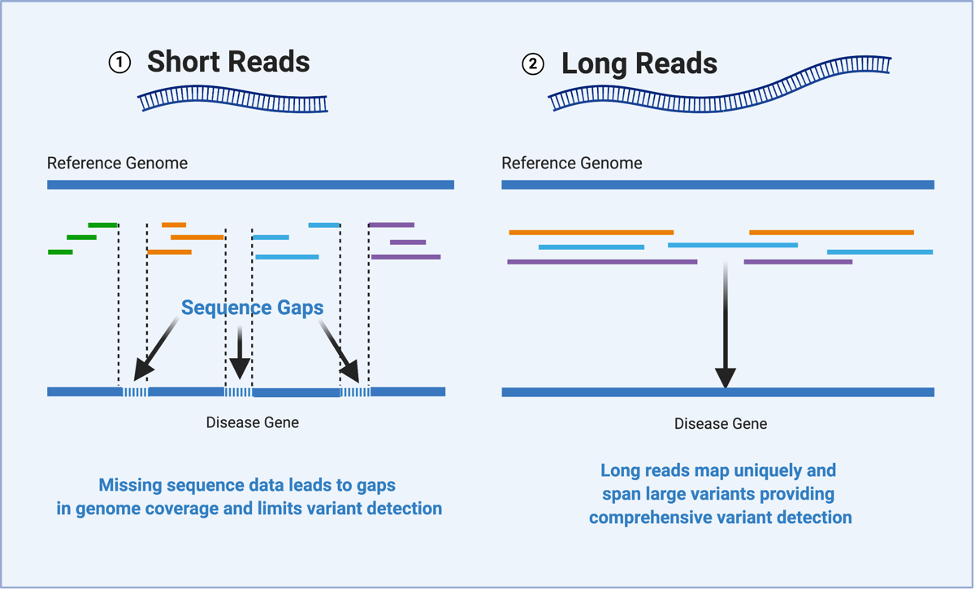
Traditionally, genome sequencing is performed by generating millions of “short” sequences, called reads, which are generally around 150 base pairs long. These short-reads are pieced back together like a puzzle using a human reference genome as a template. The reference genome is a representative example of a set of chromosomes in a human, produced from the DNA of thirteen anonymous volunteers. Genetic variants are detected by comparing the puzzle pieces to the reference genome to look for variations in the sequences.
It has been shown that despite the use of sophisticated computer technology, it is often impossible to accurately map short reads originating from some regions of DNA that have certain types of variants, like structural variants, repetitive sequences, or mobile element insertions that can move around within a DNA segment. Inaccurate or incomplete re-assembly of the DNA sequence makes it hard to detect genetic variants.
Using long-read sequencing to identify disease-causing genetic variants
Through programs like the Clinical Sequencing Evidence-Generating Research (CSER) Consortium and the Alabama Genomic Health Initiative (AGHI), HudsonAlpha Faculty Investigator Greg Cooper, PhD, and his lab have sequenced more than 1467 affected children and approximately 1511 parents. They have found genetic cause for about twenty-seven percent of the affected children, leading to a more precise and definitive clinical diagnosis.
While information gleaned from next-generation sequencing technology has led to many diagnoses, the team is constantly looking for ways to improve the technology and increase diagnostic rates for patients with rare disease. One potential approach to overcoming the limitations of short-read technology is to use a sequencing platform that produces longer reads. Recent advances in long-read sequencing now allow for the production of reads that are up to 1,000 times longer than those from short-read sequencing. Having fewer, bigger puzzle pieces leads to fewer gaps in the whole sequence once assembled. More complete coverage of the DNA sequence allows researchers and clinicians to more accurately detect variants, including those that are potentially disease-causing.
In a recent study published in Human Genetics and Genomics Advances, Cooper and his lab describe how long-read sequencing helped them identify pathogenic variants responsible for previously undiagnosable, rare neurodevelopmental disorders in two young patients. The research team, led by senior scientist and first author Susan Hiatt, PhD, used Pacific Biosciences Circular Consensus Sequencing, or “HiFi”, technology on the latest platform the PacBio Sequel 2 during the study. During HiFi sequencing, fragments of DNA are circularized and then sequenced over and over. This leads to sequence reads that are both long—clocking in at tens of thousands of bases —and accurate.
“HiFi sequencing is a cutting-edge technology that really helps us get a more accurate picture of a DNA sequence,” says Jane Grimwood, HudsonAlpha Faculty Investigator and co-director of the HudsonAlpha Genome Sequencing Center. “After we receive a DNA sample from a patient, it is sized into 15-25 thousand basepair length pieces and then sequenced. The HiFi technology reads the bases multiple times from a single molecule to produce highly accurate long-read sequences from a person’s DNA.”
Using the HiFi technology, the team analyzed six family trios (mom, dad, and child) that each had a child suspected of having a genetic neurodevelopmental disorder. Although the families had previously had their genomes sequenced with short-read technology, no disease-causing genetic variant had been identified.
The researchers found many genetic variants in each family that had previously been missed by short-read sequencing. Among these newly detected variants, the team identified likely pathogenic variation in two of the six children, meaning the DNA variant they identified is likely the cause of the child’s disease.
In one case, the team identified a likely pathogenic DNA insertion of nearly 7,000 bases in a gene called CDKL5. “Because variation in CDKL5 has been associated with early infantile epileptic encephalopathy 2, a well-characterized neurodevelopmental condition that fits well with this patient’s presentation, we dove deeper into this variant,” said Hiatt. “We performed an experiment to determine the effect of this large insertion in the CDKL5 gene, and confirmed that the gene loses its function with this insertion.”
By providing clinicians with more accurate long-read sequencing data combined with supporting data from basic biology experiments, researchers can provide a picture of a possible disease diagnosis that they did not previously have with short-read sequencing alone. Although the study looked at only six patients, it is very promising that two of the six patients now have potential answers for their undiagnosed neurodevelopmental diseases.
“The ability to find so many variants that were previously missed is exciting, and holds great promise for diagnostic testing in the future,” says Cooper. “Long-read genome sequencing will become a powerful tool for research and clinical testing over the next few years.”
Learn more about the types of DNA sequencing and how scientists are using them at http://hudsonalpha.org/piecing-together-the-genome-the-long-and-short-of-it-all.
About HudsonAlpha: HudsonAlpha Institute for Biotechnology is a nonprofit institute dedicated to developing and applying scientific advances to health, agriculture, learning, and commercialization. Opened in 2008, HudsonAlpha’s vision is to leverage the synergy between discovery, education, medicine, and economic development in genomic sciences to improve the human condition around the globe. The HudsonAlpha biotechnology campus consists of 152 acres nestled within Cummings Research Park, the nation’s second largest research park. The state-of-the-art facilities co-locate nonprofit scientific researchers with entrepreneurs and educators. HudsonAlpha has become a national and international leader in genetics and genomics research and biotech education and fosters more than 40 diverse biotech companies on campus. To learn more about HudsonAlpha, visit hudsonalpha.org.